 |
 |
| Anim Biosci > Volume 36(6); 2023 > Article |
|
Abstract
Objective
This study aims to evaluate the target genes of gga-miR-20a-5p and the regulated immune responses in the chicken macrophage cell line, HD11, by the exosome-mediated delivery of miR-20a-5p.
Methods
Exosomes were purified from the chicken macrophage cell line HD11. Then, mimic gga-miR-20p or negative control miRNA were internalized into HD11 exosomes. HD11 cells were transfected with gga-miR-20a-5p or negative control miRNA containing exosomes. After 44 h of transfection, cells were incubated with or without 5 μg/mL poly(I:C) for 4 h. Then, expression of target genes and cytokines was evaluated by quantitative real-time polymerase chain reaction.
Results
Using a luciferase reporter assay, we identified that gga-miR-20a-5p directly targeted interferon gamma receptor 2 (IFNGR2), mitogen-activated protein kinase 1 (MAPK1), mitogen-activated protein kinase kinase kinase 5 (MAP3K5), and mitogen-activated protein kinase kinase kinase 14 (MAP3K14). Moreover, the exosome-mediated delivery of gga-miR-20a-5p successfully repressed the expression of IFNGR2, MAPK1, MAP3K5, and MAP3K14 in HD11 cells. The expressions of interferon-stimulated genes (MX dynamin like GTPase 1 [MX1], eukaryotic translation initiation factor 2A [EIF2A], and oligoadenylate synthase-like [OASL]) and proinflammatory cytokines (interferon-gamma [IFNG], interleukin-1 beta [IL1B], and tumor necrosis factor-alpha [TNFA]) were also downregulated by exosomal miR-20a-5p. In addition, the proliferation of HD11 cells was increased by exosomal miR-20a-5p.
Conclusion
The exosome-mediated delivery of gga-miR-20a-5p regulated immune responses by controlling the MAPK and apoptotic signaling pathways. Furthermore, we expected that exosomal miR-20a-5p could maintain immune homeostasis against highly pathogenic avian influenza virus H5N1 infection by regulating the expression of proinflammatory cytokines and cell death.
Exosomes are cell membrane-derived small vesicles with diameters ranging from 30 to 150 nm that are generated by various cell types in biological fluids, such as plasma, lymph, saliva, semen, urine, and cerebrospinal fluid [1,2]. Exosomes contain various proteins and nucleic acids originating from “mother cells”, which are transmitted to recipient cells [3,4]. Particularly, exosomal microRNAs (miRNAs) regulate the gene expression of recipient cells by repressing and degrading their target mRNAs [5,6]. MiRNAs are small non-coding RNAs that are 18 to 23 nucleotides long that bind to the 3′-untranslated regions (UTRs) of the target mRNA complementary sequences. The miRNAs then repress and degrade the target mRNAs by forming an RNA-induced silencing complex and inhibiting mRNA translation [7]. Exosomal miRNAs derived from various cell types, such as immune cells, cancer cells, and stem cells, have been demonstrated to regulate immune responses, cell migration, metastasis, proliferation, and invasion by repressing target gene translation [8–10].
Recently, studies have been conducted on exosomes as a gene delivery vehicle for molecules, such as RNA or DNA [11–15]. Exosomes as “nature’s delivery system” have several advantages. Because exosomes are produced from host cells, they have low toxicity and immunogenicity and are easily internalized due to their exosomal host cell-derived membranes [16,17]. Moreover, exosomal membranes protect their nucleic acid contents from nuclease degradation [11].
MiR-20a, a member of the miR-17 family, is known as a regulator of immune responses [18]. For example, the expression of miR-20a-5p was also downregulated by stimulation of toll-like receptor 4 (TLR4) and TLR2 ligands in rheumatoid arthritis fibroblast-like synoviocytes [19]. Furthermore, miR-20a-5p decreased the expression of cytokines, such as interleukin (IL)-6, C-X-C motif chemokine ligand 10 (CXCL10), IL-1β, and tumor necrosis factor alpha (TNF-α), in macrophages. Moreover, the downregulated expression of miR-20a-5p by Mycobacterium tuberculosis induced the apoptosis of human macrophages by regulating c-Jun N-terminal kinase 2 (JNK2) [20]. MiR-20a-5p expression was downregulated in CD4+ T cells derived from patients with Vogt-Koyanagi-Harada disease and upregulation of miR-20a-5p suppressed the production of IL-17 by targeting oncostatin M and C-C motif chemokine ligand 1 [21].
In our previous study, the expression of gga-miR-20a-5p was down-regulated in exosomes derived from highly pathogenic avian influenza virus (HPAIV) H5N1-infected chickens compared with exosomes of non-infected chickens, and 32 immune-related genes were predicted as gga-miR-20a-5p targets [22]. However, there are limited studies about the regulatory function of miR-20a-5p in chickens. Therefore, in this study, we evaluated the target genes of gga-miR-20a-5p and demonstrated the immunomodulatory effects in a chicken macrophage cell line by the exosome-mediated delivery of miR-20a-5p.
The chicken macrophage cell line HD11 [23] was maintained at Roswell Park Memorial Institute (RPMI) in 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 100 IU/mL penicillin, 100 mg/mL streptomycin, and 10% fetal bovine serum (Thermo Fisher Scientific, USA) in a humidified incubator with 5% CO2 at 41°C. To purify exosomes, HD11 cells (1.0×107) were seeded in three 100-mm cell culture dishes (SPL Life Sciences, Pocheon, Korea) in exosome-depleted RPMI 1640 medium containing 100 IU/mL penicillin, 100 mg/mL streptomycin, and 10% exosome-depleted fetal bovine serum (#EXO-FBSHI-250A-1; System Bioscience, Palo Alto, CA, USA) and incubated for 24 h. The cell culture supernatant was collected to purify exosomes using the ExoQuick-TC kit (#EXOTC50A-1; System Biosciences, USA) according to the manufacturer’s protocol. The concentration of the purified exosomes was measured using a Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. For the characterization of exosomes, their particle size was measured using a nanoparticle analyzer (SZ-100; Horiba, Kyoto, Japan). Furthermore, western blotting was performed using antibodies against CD9 (#13174; Cell Signaling Technology, Danvers, MA, USA) and CD81 (#56039; Cell Signaling Technology, USA), according to previously described methods [24].
We loaded miRNA to exosomes as described by Kim et al [25] with a slight modification. Mimic gga-miR-20a-5p and miR-NC (negative control miRNA) were synthesized by Bioneer (Daejeon, Korea). The sequence of the mimic gga-miR-20a-5p was 5′-UAAAGUGCUUAUAGUGCAG GUAG-3′ and that of the miR-NC was 5′-UUCUCCGAA CGUGUCACGU-3′. For loading of the miRNAs into the exosomes, 20 pmol of mimic gga-miR-20-5p or miR-NC were diluted in 100 μL of Opti-MEM I Reduced Serum Medium (Gibco, Waltham, MA, USA) and 6 μL of Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, San Diego, CA, USA) was diluted in 100 μL of Opti-MEM I Reduced Serum Medium. Diluted miRNA solutions were added to the diluted Lipofectamine RNAiMAX and incubated for 20 min at room temperature. Next, 100 μg of purified exosomes were incubated with miRNA-Lipofectamine RNAiMAX complexes for 6 h in a humidified incubator with 5% CO2 at 41°C. Then, the complexes were filtered with a 100 kDa Amicon Ultra-0.5 Centrifugal Filter (Merck Millipore, Burlington, MA, USA) to remove un-loaded miRNA in the exosomes. Afterward, 1.2×106 cells/well of HD11 cells were plated in 12-well plates (SPL, Korea) containing 1.0 mL of exosome-depleted RPMI 1640 medium and incubated with the concentrated exosomes. After 44 h of incubation, cells were incubated with or without 5 μg/mL of polyinosine-polycytidylic acid (poly[I:C]) high molecular weight (HMW) (Invitrogen, USA) for 4 h. Then, the total RNA of the HD11 cells was extracted using TRIzol reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. The cDNA was then synthesized using 2 μg of RNA with the RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol.
Primers for quantitative real-time polymerase chain reaction (qRT-PCR) were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primerblast/) and synthesized by Genotech (Daejeon, Korea) (Table 1). qRT-PCR was performed using Dyne qPCR 2X PreMIX (Dyne Bio, Seongnam, Korea) according to the manufacturer’s protocol with the CFX Connect Real-Time PCR Detection System (Bio-rad, Hercules, CA, USA). Chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for internal reference. The relative quantification of mRNA expression levels was calculated using the 2–ΔΔCt method [26].
To quantify miRNA expression, cDNA was synthesized using 2 μg of RNA with the Mir-X miRNA First-Strand Synthesis Kit (TAKARA Bio Inc., Otsu, Shiga, Japan) according to the manufacturer’s protocol. The forward primers of gga-miR-20a-5p, 5′-TAAAGTGCTTATAGTGCAGGTAG -3′, and small nuclear ribonucleoprotein polypeptide A (U1A), 5′-CTGCATAATTTGTGGTAGTGG -3′, were synthesized by Genotech (Korea). The miRNA expression level was evaluated using the Mir-X miRNA qRT-PCR TB Green Kit (TAKARA Bio Inc., Japan) with the CFX Connect Real-Time PCR Detection System (Bio-rad, USA). The miRNA expression level was calculated using the 2–ΔΔCt method with U1A for internal reference.
For vector construction, the 3′-UTR of chicken interferon gamma receptor 2 (IFNGR2, NM_001008676), mitogen-activated protein kinase 1 (MAPK1, XM_015275131), mitogen-activated protein kinase kinase kinase 5 (MAP3K5, XM_015 284184), and mitogen-activated protein kinase kinase kinase 14 (MAP3K14, NM_001030927), including the binding site of miR-20a-5p, were amplified using wild type (WT) primers and mutant (Mut) primers (Table 1). The amplified PCR products of WT- and Mut-IFNGR2, MAPK1, MAP3K5, and MAP3K14 were cloned into a pMIR-REPORT luciferase vector (Thermo Fisher Scientific, USA). For the luciferase reporter assay, 1.2×106 cells/well of the chicken fibroblast cell line DF-1 were seeded in a 6-well plate (SPL, Korea) containing 2.0 mL of culture medium and co-transfected with 40 pmol of miR-20-5p or miR-NC, 900 ng of WT or Mut plasmids, and 100 ng of the pMIR-REPORT β-gal control vector using the Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. After 24 h of transfection, luciferase activity was detected using the Luciferase Assay System (Promega, Madison, WA, USA) and β-galactosidase activity was detected using the β-galactosidase enzyme assay system (Promega, USA) for normalization of transfection efficiency.
HD11 cells (6.0×105 cells/well) were plated in 24-well plates (SPL, Korea) containing 0.5 mL of exosome-depleted RPMI 1640 medium and transfected with exosomal gga-miR-20a-5p or miR-NC according to the method described above. After 44 h of transfection, cells were incubated with or without 5 μg/mL of poly(I:C) HMW (Invivogen, San Diego, CA, USA) for 4 h. Then, cell proliferation was measured using the Cell Counting Kit-8 (Enzo Life Science, Oyster Bay, NY, USA) according to the manufacturer’s protocol.
Data are presented as the mean±standard error of the mean of three independent experiments. Statistical analyses were performed using the IBM SPSS software (SPSS 26.0; IBM, Chicago, IL, USA). Statistical comparison between two groups were evaluated using Student’s t-test. Statistical significance is defined when p-values are less than 0.05.
The gga-miR-20a-5p target genes were predicted using miRDB ( http://mirdb.org/) and TargetScan ( https://www.targetscan.org/vert_80/). Then, IFNGR2, MAPK1, MAP3K5, and MAP3K14 genes were selected as candidate targets of gga-miR-20a-5p. To validate the target genes, a luciferase reporter assay was conducted by transfecting DF-1 cells with the WT or Mut 3′-UTR of IFNGR2, MAPK1, MAP3K5, or MAP3K14, along with mimic gga-miR-20a-5p or miR-NC. The luciferase activity of the cells transfected with WT IFNGR2, MAPK1, MAP3K5, and MAP3K14 was significantly reduced by miR-20a-5p compared with miR-NC, but the luciferase activity of the cells transfected with Mut IFNGR2, MAPK1, MAP3K5, and MAP3K14 was not reduced by miR-20a-5p (Figure 2).
After transfection of the mimic gga-miR-20a-5p into the HD11 cells using exosomes, the transfection efficiency was evaluated by qRT-PCR (Figure 3). The expression of miR-20a-5p was significantly increased by transfection of miR-20a-5p using exosomes. Moreover, the expression of IFNGR2, MAPK1, MAP3K5, and MAP3K14 was evaluated (Figure 4). The expressions of IFNGR2 and MAP3K14 were significantly downregulated in HD11 cells transfected by exosomal miR-20a-5p and poly(I:C)-stimulated exosomal miR-20a-5p-transfected HD11 cells. The expressions of MAPK1 and MAP3K5 were significantly downregulated in only poly(I:C)-stimulated HD11 cells transfected with exosomal miR-20a-5p.
Because interferon (IFN)-γ induces interferon-stimulated gene (ISG) transcription and stimulates the expression of proinflammatory cytokines through the MAPK signaling pathway, the expression of ISGs and proinflammatory cytokines was evaluated by qRT-PCR (Figure 5). The expressions of ISGs, such as MX dynamin like GTPase 1 (MX1), eukaryotic translation initiation factor 2A (EIF2A), and oligoadenylate synthase-like (OASL) were downregulated in poly(I:C)-stimulated HD11 cells transfected by exosomal miR-20a-5p compared to cells transfected by exosomal miR-NC. Moreover, the expressions of proinflammatory cytokines, such as IFN-γ, IL-1β, and TNF-α, were downregulated in poly(I:C)-stimulated HD11 cells transfected by exosomal miR-20a-5p compared to those of the poly(I:C)-stimulated cells transfected by exosomal miR-NC.
Since MAP3K5 and MAP3K14 are signaling molecules of apoptosis, we measured cell proliferation after exosome-mediated transfection of miR-20a-5p. Cell proliferation was increased by transfection with exosomal miR-20a-5p only and exosomal miR-20a-5p with poly(I:C) stimulation (Figure 6).
In the present study, we delivered gga-miR-20a-5p via exosomes derived from a chicken macrophage cell line and identified the gene regulatory functions of gga-miR-20a-5p, which repressed the expression of IFNGR2, MAPK1, MAP3K5, and MAP3K14, in chicken macrophage cells.
MiR-20a-5p is a known regulator of immune responses. For example, miR-20a-5p was downregulated during liver fibrosis and transfection of miR-20a-5p reduced the production of the proinflammatory cytokines IL-6, TNF-α, IL-18, and IFN-γ in a murine hepatoma cell line by targeting transforming growth factor beta 2 (TGFB2) [27]. MiR-20a-5p is also upregulated in certain cancers, such as thyroid cancer [28], gastric cancer [29], breast cancer [30], and cervical cancer [31], by inducing cancer cell proliferation and inhibiting cell apoptosis. On the other hand, miR-20a-5p is normally downregulated in infectious and proinflammatory environments, such as tuberculosis [20], liver fibrosis [27], and rheumatoid arthritis [19], by stimulating proinflammatory cytokine production and apoptosis. In our previous study, the expression of exosomal miR-20a-5p was downregulated in H5N1-infected chickens [22]. Therefore, we predicted the target genes and validated the regulatory functions in chicken macrophages.
In this study, we stimulated chicken macrophages with poly(I:C) after transfection with exosomal miR-20a-5p. Macrophages as antigen presenting cells have crucial immune functions, such as phagocytosis of pathogens and production of cytokines and chemokines [32]. Moreover, poly(I:C) is a viral like double-stranded RNA (dsRNA) that binds to TLR3, RNA helicases, retinoic acid-inducible gene-I, and melanoma differentiation-associated protein 5. Upon recognition, the transcription of pro-inflammatory cytokines and interferons are activated by the transcription factors such as interferon regulatory factor-3 (IRF-3), active protein-1 (AP-1), and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) [33,34].
IFNGR2, MAPK1, MAP3K5, and MAP3K14 were predicted as candidate target genes of gga-miR-20a-5p. In the canonical signaling pathway, after binding of IFN-γ and its receptors (IFNGR1 and IFNGR2), phosphorylation of signal transducer and activator of transcription 1 (STAT1) by the Janus kinase 1 (JAK1) and Janus kinase 2 (JAK2) proteins induces STAT1 dimerization and translocation into the nucleus to activate ISG transcription [35]. In the non-canonical signaling pathway, IFN-γ activates various molecules, such as extracellular signal-regulated kinase (ERK), phosphoinositide 3-kinase (PI3K), proline-rich tyrosine kinase 2 (Pyk2), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), and ribonuclease A family 1 (Raf1). [36]. In this study, we identified that exosomal miR-20a-5p repressed the expression of IFNGR2 and ISGs, such as MX1, EIF2A, and OASL, were downregulated in poly(I:C)-stimulated HD11 cells by exosomal miR-20a-5p. Type II IFNs, such as IFN-γ stimulate the phosphorylation of STAT1 by binding to IFNGR1 and IFNGR2. Then, gamma-activated sequences are activated by phosphorylated STAT1 and proinflammatory genes such as IL-1β and ISGs are induced [37]. Therefore, we suggest that the repressed expression of IFNGR2 by exosomal miR-20a-5p decreased ISG transcription following the inhibited IFN-γ signal. The other candidate target genes, MAPK1 (ERK2), MAP3K5 (ASK1), and MAP3K14 (NIK), are MAPK signaling pathway molecules (KEGG ID; gga04010). Moreover, MAP3K5 and MAP3K14 are also related to the apoptosis pathway (KEGG ID; gga04210). The TLR3 signaling pathway is known to activate the MAPK signaling pathway by inducing proinflammatory cytokine expression [38,39] and apoptosis [40]. Furthermore, the expression of TNF-α, a critical inflammatory cytokine responsible for necrosis or apoptosis, is also stimulated by the TLR3 signaling pathway [41,42]. In this study, we demonstrated that the expression of MAPK1, MAP3K5, and MAP3K14 was downregulated by exosomal miR-20a-5p (Figures 2 and 4). In addition, the proliferation of chicken macrophage cells was increased by transfection of exosomal miR-20a-5p only and exosomal miR-20a-5p with poly(I:C) stimulation compared with transfection using exosomal miR-NC (Figure 6). Moreover, the expression of TNF-α was downregulated in poly(I:C)-stimulated macrophages by transfection of exosomal miR-20a-5p (Figure 5). Therefore, we suggest that the repressed expression of the MAPK signaling pathway molecules, MAPK1, MAP3K5, and MAP3K14, reduced TNF-α expression. Moreover, along with TNF-α reduction, the repressed expression of the apoptotic pathway molecules, MAP3K5 and MAP3K14, increased the proliferation of chicken macrophages.
HPAIV H5N1 infection induces a massive production of proinflammatory cytokines, called a cytokine storm, which causes multiple organ dysfunction syndrome, acute respiratory distress syndrome, and sudden death [43–46]. Moreover, the MAPK signaling pathway is an important mediator of cytokine production against influenza A virus infection [47]. Therefore, we expected that the exosome-mediated delivery of gga-miR-20a-5p to H5N1-infected chicken cells could regulate proinflammatory cytokine expression by targeting the MAPK signaling pathway molecules, MAPK1, MAP3K5, and MAP3K14, and inhibit cell death by targeting the apoptosis signaling pathway molecules, MAP3K5 and MAP3K14.
In the present study, we evaluated gga-miR-20a-5p functions by delivery of exosomes into chicken macrophage cell lines. The expression of the candidate target genes, IFNGR2, MAPK1, MAP3K5, and MAP3K14, was repressed by exosomal miR-20a-5p. Moreover, the expression of ISGs and proinflammatory cytokines was downregulated by exosomal miR-20a-5p targeting IFNGR2 and MAPK signaling pathway molecules (MAPK1, MAP3K5, and MAP3K14). Cell proliferation was increased by the exosomal miR-20a-5p targeting the apoptosis pathway molecules (MAP3K5 and MAP3K14). However, although we identified repressed expression of miR-20a-5p target genes via a luciferase reporter assay and qRT-PCR, protein expression levels of the target genes were not measured because there are no commercially available antibodies against these chicken proteins. Moreover, additional studies such as evaluation of macrophage phenotypic changes and antiviral activity would be helpful in clarifying these results and conclusions, so continuous research on exosomal miR-20a-5p is needed. Taken together, the exosome-mediated delivery of gga-miR-20a-5p will help to maintain immune homeostasis against virus infections like HPAIV H5N1 by repressing the expression of proinflammatory cytokines and cell death.
Notes
AUTHOR CONTRIBUTIONS
YH and YHH conceived and designed the experiments. YH, THV, JH, and SK performed the experiments. YH analyzed the data. YH and YHH wrote the manuscript. HSL and YHH reviewed and revised the paper. All authors read and approved the final manuscript.
Figure 1
Characterization of exosomes. (A) Particle size distribution of exosomes in the chicken macrophage cell line HD11 measured using a nanoparticle analyzer. (B) Detection of exosomal markers, CD9 and CD81, by western blotting.
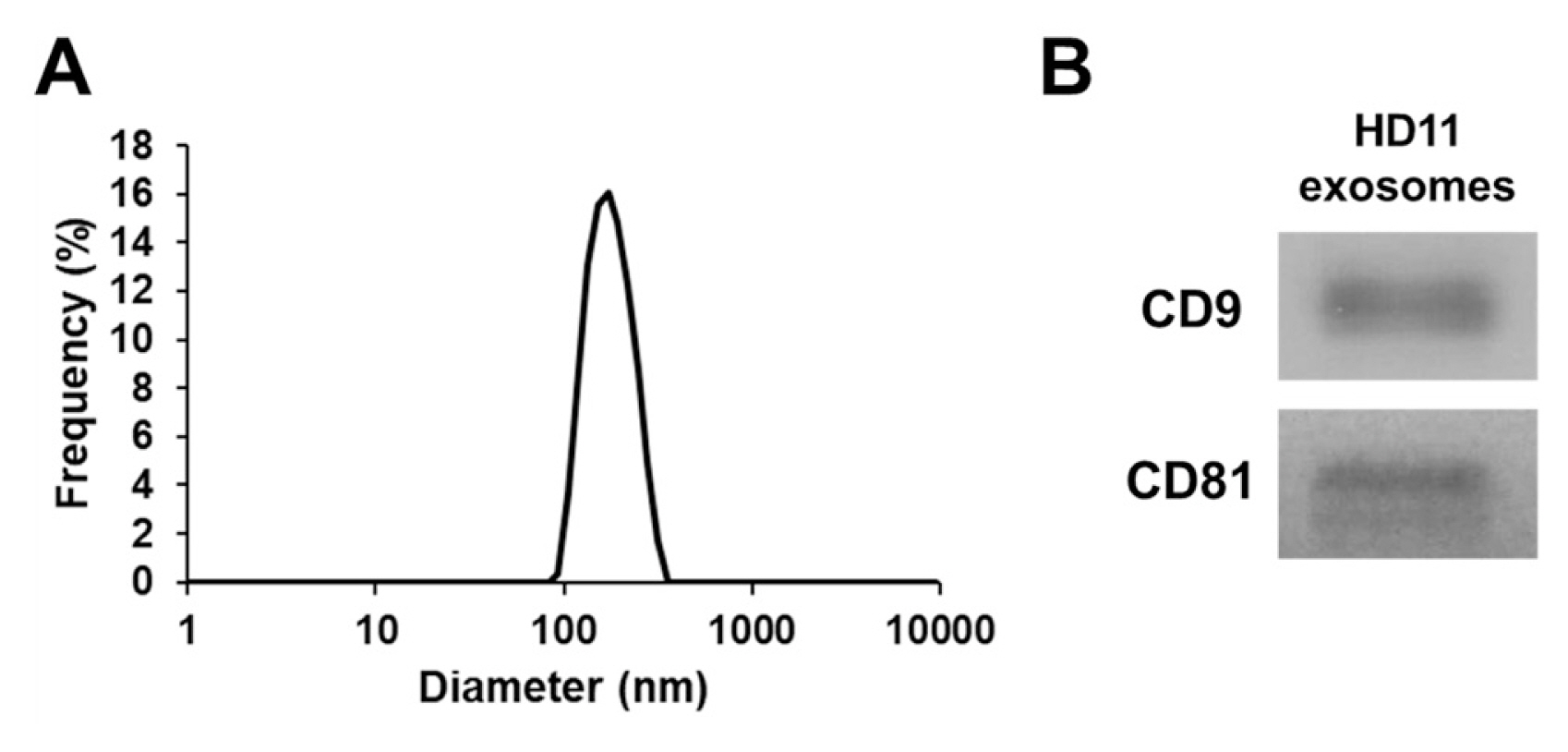
Figure 2
Luciferase reporter assay and potential binding site of gga-miR-20a-5p in target genes. For the luciferase reporter assay, the wild type (WT) or mutant (Mut) 3′-untranslated region (UTR) of (A) IFNGR2, (B) MAPK1, (C) MAP3K5, and (D) MAP3K14 were cloned into pMIR-REPORT luciferase vectors and co-transfected with mimic miR-20a-5p or negative control microRNA (miRNA) (miR-NC) in DF-1 cells by lipofectamine. IFNGR2, interferon gamma receptor 2; MAPK1, mitogen-activated protein kinase 1; MAP3K5, mitogen-activated protein kinase kinase kinase 5; MAP3K14, mitogen-activated protein kinase kinase kinase 14. * p<0.05 and *** p<0.001.
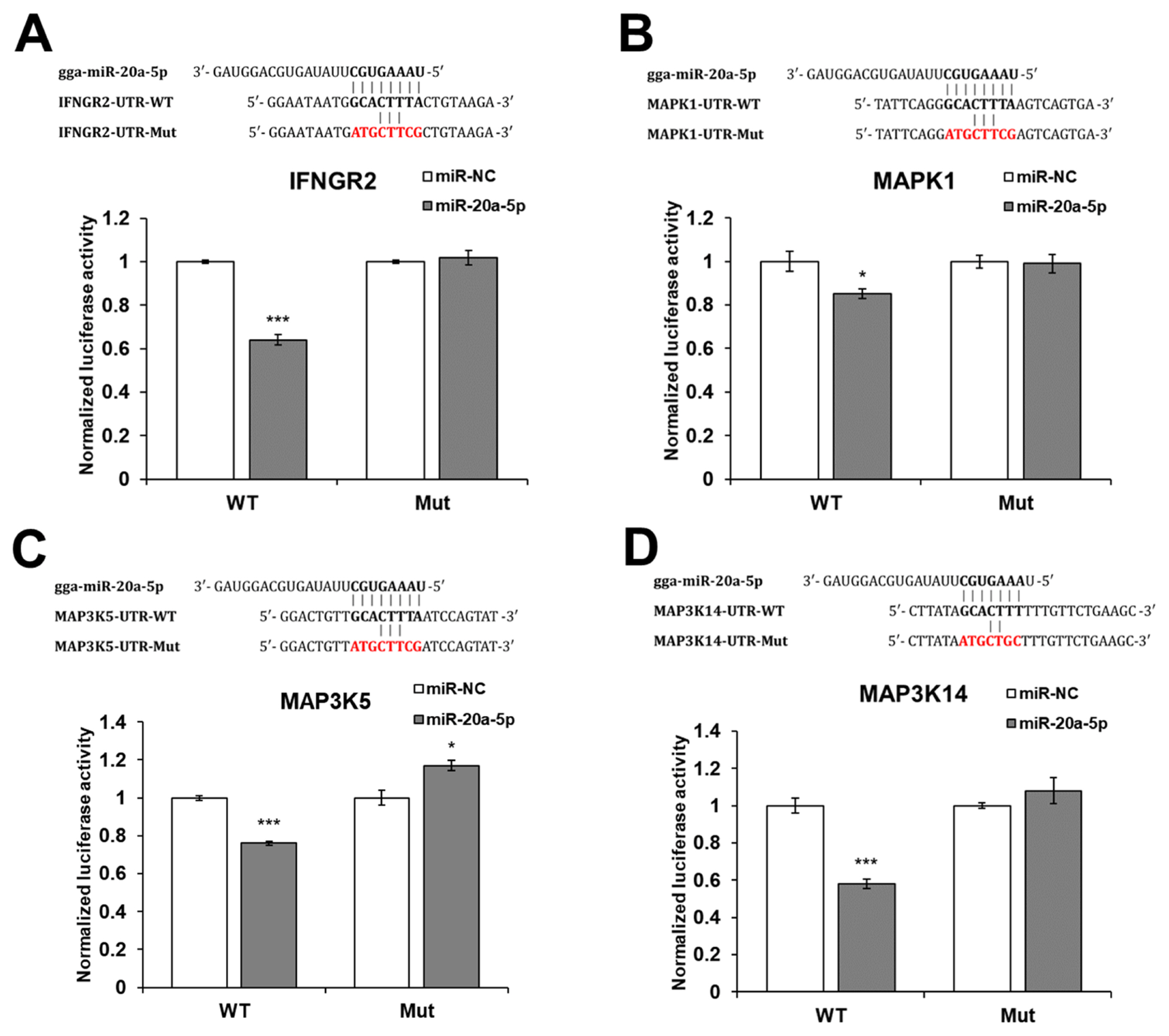
Figure 3
MicroRNA (miRNA) delivery efficiency using exosomes. HD11 cells were transfected with exosomes containing gga-miR-20a-5p or negative control miRNA (miR-NC). Then, miRNA delivery efficiency was evaluated by quantitative real-time polymerase chain reaction (qRT-PCR). Relative quantitation data are represented as mean±standard error of the mean (SEM) normalized to U1A using the 2−ΔΔCt method. Data are expressed as mean±SEM of three independent experiments. *** p<0.001.
Figure 4
Exosome-mediated delivery of miR-20a-5p repressed target genes in vitro. HD11 cells were transfected with gga-miR-20a-5p or negative control microRNA (miRNA) (miR-NC) containing exosomes. After 44 h of transfection, cells were incubated with or without 5 μg/mL poly(I:C) for 4 h. Then, expression of the target genes, IFNGR2, MAPK1, MAP3K5, and MAP3K14, was evaluated by quantitative real-time polymerase chain reaction (qRT-PCR). Data are expressed as the mean±standard error of the mean (SEM) and are representative of three independent experiments. IFNGR2, interferon gamma receptor 2; MAPK1, mitogen-activated protein kinase 1; MAP3K5, mitogen-activated protein kinase kinase kinase 5; MAP3K14, mitogen-activated protein kinase kinase kinase 14. * p<0.05, ** p<0.01, *** p<0.001.
Figure 5
Exosome-mediated delivery of miR-20a-5p downregulated the expression of interferon-stimulated genes and proinflammatory cytokines. HD11 cells were transfected with gga-miR-20a-5p or negative control microRNA (miRNA) (miR-NC) containing exosomes. After 44 h of transfection, cells were incubated with or without 5 μg/mL of poly(I:C) for 4 h. Then, expressions of interferon stimulated genes (MX1, EIF2A, and OASL) and proinflammatory cytokines (IFN-γ, IL-1β, and TNF-α) were evaluated by quantitative real-time polymerase chain reaction (qRT-PCR). Data are expressed as the mean±standard error of the mean (SEM) and are representative of three independent experiments. MX1, MX dynamin like GTPase 1; EIF2A, eukaryotic translation initiation factor 2A; OASL, oligoadenylate synthase-like; IFN-γ, interferon gamma; IL-1β, interleukin-1beta; TNF-α, tumor necrosis factor alpha. * p<0.05 and *** p<0.001.

Figure 6
Exosome-mediated delivery of miR-20a-5p increased cell proliferation. HD11 cells were transfected with gga-miR-20a-5p or negative control microRNA (miRNA) (miR-NC) containing exosomes. After 44 h of transfection, cells were incubated with or without 5 μg/mL of poly(I:C) for 4 h. Then, cell proliferation was measured by the Cell Counting Kit-8 assay. Data are expressed as the mean±standard error of the mean and are representative of three independent experiments. * p<0.05 and ** p<0.01.
Table 1
Sequences of primers used for quantitative real-time polymerase chain reaction analysis
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IFNGR2, interferon gamma receptor 2; MAPK1, mitogen-activated protein kinase 1; MAP3K5, mitogen-activated protein kinase kinase kinase 5; MAP3K14, mitogen-activated protein kinase kinase kinase 14; MX1, MX dynamin like GTPase 1; EIF2AK2, eukaryotic translation initiation factor 2 alpha kinase 2; OASL, oligoadenylate synthase-like; IFNG, interferon gamma; IL1B, interleukin-1beta; TNFA, tumor necrosis factor alpha.
REFERENCES
1. Lu M, Xing H, Xun Z, et al. Exosome-based small RNA delivery: progress and prospects. Asian J Pharm 2018; 13:1–11. https://doi.org/10.1016/j.ajps.2017.07.008



2. Bakhshandeh B, Amin Kamaleddin M, Aalishah K. A comprehensive review on exosomes and microvesicles as epigenetic factors. Curr Stem Cell Res Ther 2017; 12:31–6. https://doi.org/10.2174/1574888X11666160709211528


3. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010; 73:1907–20. https://doi.org/10.1016/j.jprot.2010.06.006


4. Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol 2012; 14:1036–45. https://doi.org/10.1038/ncb2574


5. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell 2016; 164:1226–32. https://doi.org/10.1016/j.cell.2016.01.043


6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97. https://doi.org/10.1016/S0092-8674(04)00045-5


7. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522–31. https://doi.org/10.1038/nrg1379


8. Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014; 26:707–21. https://doi.org/10.1016/j.ccell.2014.09.005



9. Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci 2013; 14:14240–69. https://doi.org/10.3390/ijms140714240



10. Li XJ, Ren ZJ, Tang JH, Yu Q. Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol Biochem 2017; 44:1741–8. https://doi.org/10.1159/000485780


11. Jeong K, Yu YJ, You JY, Rhee WJ, Kim JA. Exosome-mediated microrna-497 delivery for anti-cancer therapy in a microfluidic 3d lung cancer model. Lab Chip 2020; 20:548–57. https://doi.org/10.1039/c9lc00958b


12. Lee GW, Thangavelu M, Choi MJ, et al. Exosome mediated transfer of miRNA-140 promotes enhanced chondrogenic differentiation of bone marrow stem cells for enhanced cartilage repair and regeneration. J Cell Biochem 2020; 121:3642–52. https://doi.org/10.1002/jcb.29657


13. Rezaei R, Baghaei K, Amani D, et al. Exosome-mediated delivery of functionally active miRNA-375-3p mimic regulate epithelial mesenchymal transition (EMT) of colon cancer cells. Life Sci 2021; 269:119035https://doi.org/10.1016/j.lfs.2021.119035


14. Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles—endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta Rev Cancer 2014; 1846:75–87. https://doi.org/10.1016/j.bbcan.2014.04.005

15. Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol Pharmaceutics 2015; 12:3650–7. https://doi.org/10.1021/acs.molpharmaceut.5b00364



16. Paranjpe M, Müller-Goymann CC. Nanoparticle-mediated pulmonary drug delivery: a review. Int J Mol Sci 2014; 15:5852–73. https://doi.org/10.3390/ijms15045852



17. Tan S, Wu T, Zhang D, Zhang Z. Cell or cell membrane-based drug delivery systems. Theranostics 2015; 5:863–81. https://doi.org/10.7150/thno.11852



18. Wu Y, Liu Y, Wang Q, et al. MiR-20a-5p is multifunctional regulator in chickens immune responses induced by NDV, IBDV and AIV vaccines respectively. Res Sq. 2021. Nov. 10[preprint]. https://doi.org/10.21203/rs.3.rs-1025677/v1

19. Philippe L, Alsaleh G, Pichot A, et al. MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann Rheum Dis 2013; 72:1071–9. https://doi.org/10.1136/annrheumdis-2012-201654


20. Zhang G, Liu X, Wang W, et al. Down-regulation of miR-20a-5p triggers cell apoptosis to facilitate mycobacterial clearance through targeting JNK2 in human macrophages. Cell Cycle 2016; 15:2527–38. https://doi.org/10.1080/15384101.2016.1215386



21. Chang R, Yi S, Tan X, et al. MicroRNA-20a-5p suppresses IL-17 production by targeting OSM and CCL1 in patients with Vogt-Koyanagi-Harada disease. Br J Ophthalmol 2018; 102:282–90. https://doi.org/10.1136/bjophthalmol-2017-311079


22. Hong Y, Truong AD, Lee J, et al. Exosomal miRNA profiling from H5N1 avian influenza virus-infected chickens. Vet Res 2021; 52:36https://doi.org/10.1186/s13567-021-00892-3



23. Klasing KC, Peng RK. Influence of cell sources, stimulating agents, and incubation conditions on release of interleukin-1 from chicken macrophages. Dev Comp Immunol 1987; 11:385–94. https://doi.org/10.1016/0145-305X(87)90082-6


24. Hong Y, Lee J, Vu TH, Lee S, Lillehoj HS, Hong YH. Immunomodulatory effects of avian β-defensin 5 in chicken macrophage cell line. Res Vet Sci 2020; 132:81–7. https://doi.org/10.1016/j.rvsc.2020.06.002


25. Kim H, Rhee WJ. Exosome-mediated let7c-5p delivery for breast cancer therapeutic development. Biotechnol Bioprocess Eng 2020; 25:513–20. https://doi.org/10.1007/s12257-020-0002-0

26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001; 25:402–8. https://doi.org/10.1006/meth.2001.1262


27. Fu X, Qie J, Fu Q, Chen J, Jin Y, Ding Z. miR-20a-5p/TGFBR2 axis affects pro-inflammatory macrophages and aggravates liver fibrosis. Front Oncol 2020; 10:107https://doi.org/10.3389/fonc.2020.00107



28. Xiong Y, Zhang L, Kebebew E. MiR-20a is upregulated in anaplastic thyroid cancer and targets LIMK1. PLoS One 2014; 9:e96103https://doi.org/10.1371/journal.pone.0096103



29. Wang M, Gu H, Qian H, et al. miR-17-5p/20a are important markers for gastric cancer and murine double minute 2 participates in their functional regulation. Eur J Cancer 2013; 49:2010–21. https://doi.org/10.1016/j.ejca.2012.12.017


30. Guo L, Zhu Y, Li L, et al. Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med 2019; 8:5687–701. https://doi.org/10.1002/cam4.2454



31. Laengsri V, Kerdpin U, Plabplueng C, Treeratanapiboon L, Nuchnoi P. Cervical cancer markers: epigenetics and microRNAs. Lab Med 2018; 49:97–111. https://doi.org/10.1093/labmed/lmx080


32. Hirayama D, Iida T, Nakase H. The phagocytic function of macrophage-enforcing innate immunity and tissue homeostasis. Int J Mol Sci 2018; 19:92https://doi.org/10.3390/ijms19010092



33. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by toll-like receptor 3. Nature 2001; 413:732–8. https://doi.org/10.1038/35099560


34. Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann NY Acad Sci 2008; 1143:1–20. https://doi.org/10.1196/annals.1443.020


35. Jorgovanovic D, Song M, Wang L, Zhang Y. Roles of IFN-γ in tumor progression and regression: a review. Biomark Res 2020; 8:49https://doi.org/10.1186/s40364-020-00228-x



36. Tang M, Tian L, Luo G, Yu X. Interferon-gamma-mediated osteoimmunology. Front Immunol 2018; 9:1508https://doi.org/10.3389/fimmu.2018.01508



37. Kim YM, Shin EC. Type I and III interferon responses in SARS-CoV-2 infection. Exp Mol Med 2021; 53:750–60. https://doi.org/10.1038/s12276-021-00592-0



38. Johnsen IB, Nguyen TT, Bergstrøm B, Lien E, Anthonsen MW. Toll-like receptor 3-elicited MAPK activation induces stabilization of interferon-β mRNA. Cytokine 2012; 57:337–46. https://doi.org/10.1016/j.cyto.2011.11.024


39. Peroval MY, Boyd AC, Young JR, Smith AL. A critical role for MAPK signalling pathways in the transcriptional regulation of toll like receptors. PloS One 2013; 8:e51243https://doi.org/10.1371/journal.pone.0051243



40. Sun R, Zhang Y, Lv Q, et al. Toll-like receptor 3 (TLR3) induces apoptosis via death receptors and mitochondria by up-regulating the transactivating p63 isoform α (TAP63α). J Biol Chem 2011; 286:15918–28. https://doi.org/10.1074/jbc.M110.178798



41. Richer MJ, Lavallee DJ, Shanina I, Horwitz MS. Toll-like receptor 3 signaling on macrophages is required for survival following coxsackievirus B4 infection. PloS One 2009; 4:e4127https://doi.org/10.1371/journal.pone.0004127



42. Idriss HT, Naismith JH. TNFα and the TNF receptor superfamily: Structure-function relationship (s). Microsc Res Tech 2000; 50:184–95. https://doi.org/10.1002/1097-0029(20000801)50:3<184::AID-JEMT2>3.0.CO;2-H


43. Liu Q, Zhou YH, Yang ZQ. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol 2016; 13:3–10. https://doi.org/10.1038/cmi.2015.74



44. Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev 2012; 76:16–32. https://doi.org/10.1128/MMBR.05015-11



45. D'Elia RV, Harrison K, Oyston PC, Lukaszewski RA, Clark GC. Targeting the “cytokine storm” for therapeutic benefit. Clin Vaccine Immunol 2013; 20:319–27. https://doi.org/10.1128/cvi.00636-12



46. Gu Y, Zuo X, Zhang S, et al. The mechanism behind influenza virus cytokine storm. Viruses 2021; 13:1362https://doi.org/10.3390/v13071362



47. Yu J, Sun X, Goie JYG, Zhang Y. Regulation of host immune responses against influenza a virus infection by mitogen-activated protein kinases (MAPKs). Microorganisms 2020; 8:1067https://doi.org/10.3390/microorganisms8071067



- TOOLS






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





